Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology* RIKEN Harima Insutitute/SPring-8, Japan** Iwaki-Meisei University, Japan***
○Keita Kikuchi* Hiroshi Kondo* Ella C.M. Juan* Wataru Adachi* Ryoji Masui** Seiki Kuramitsu** Kaoru Suzuki*** Takeshi Sekiguchi*** Akio Takenaka*
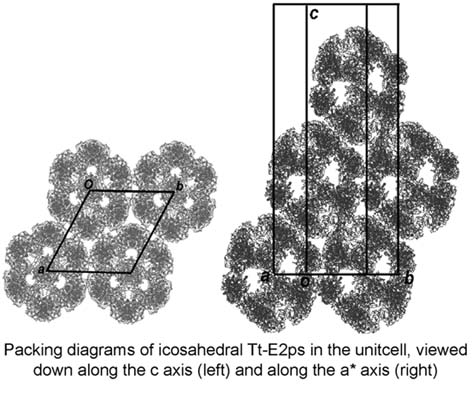
Pyruvate dehydrogenase complex (PDC) is composed of multiple copies of three enzymes E1, E2 and E3, in which E2s form the central core. There are two types of the architectures depending on organisms; one is composed of 60 subunits of E2 (with 532 symmetry), and the other of 24 subunits (with 432 symmetry). In this study, ultracentrifugation experiments were performed at first to clarify the architecture of E2 from Thermus thermophilus (Tt-E2). The resulting sedimentation coefficient corresponds to 60 copies of Tt-E2, suggesting that the core structure is similar to those of eukaryotes and Gram-positive prokaryotes despite Thermus thermophilus being Gram-negative bacteria. To determine the structure, Tt-E2 was crystallized by the hanging-drop vapor diffusion method. X-Ray diffraction data at 11 Å resolution were collected with synchrotron radiation at PF. The initial phases were successfully solved by molecular replacement assuming 532 symmetry. The crystal structure was refined with ncs constraints. The unit cell has the following dimensions, a=b=215 Å and c=543 Å with the space group R32. As shown in the figure, the icosahedral Tt-E2s are reasonably packed in the unit cell.
